Evaluating Probabilities
Published:
This blog post covers the evaluation of probabilities and probability densities for continuous variables. The motivation for this discussion came from seeing the disparity in various modeling approaches. My hope is that this blog post serves as a resource, particularly for those entering the field, on the different approaches that exist.
probability
To start, let’s first review the concept of probability. Under the Bayesian interpretation, probability, $p(X)$ expresses a degree of belief regarding the state, $X=x$, of some variable, $X$. Variables can take many different forms: binary, discrete, continuous, categorical, etc. over different spaces. For instance, a light switch can be represented as a binary variable that can take either an ‘off’ $(0)$ or ‘on’ $(1)$ state, $X \in {0, 1 }$. If we add a dimmer to the light switch, we could instead represent the output as a continuous variable over an interval from minimum to maximum brightness, $X \in [ x_\text{min}, x_\text{max} ]$.
Before observing the state of the light switch, we can express our belief about its state using probability. With the binary light switch, we might say that there is a probability of $0.9$ that the switch is in the ‘off’ state. Mathematically, we can assign a Bernoulli probability distribution to the variable, with
\[P(X=0) = 0.9\]Because probabilities must sum to $1$ (the variable must be in some state), we therefore also express the belief that the light is ‘on’ with probability
\[P(X=1) = 1 - 0.9 = 0.1.\]We have assigned a probability mass of $0.9$ to ‘off’ and $0.1$ to ‘on’. As we see, this probability mass function is just a function that assign a certain amount of mass to each state that the variable can take.
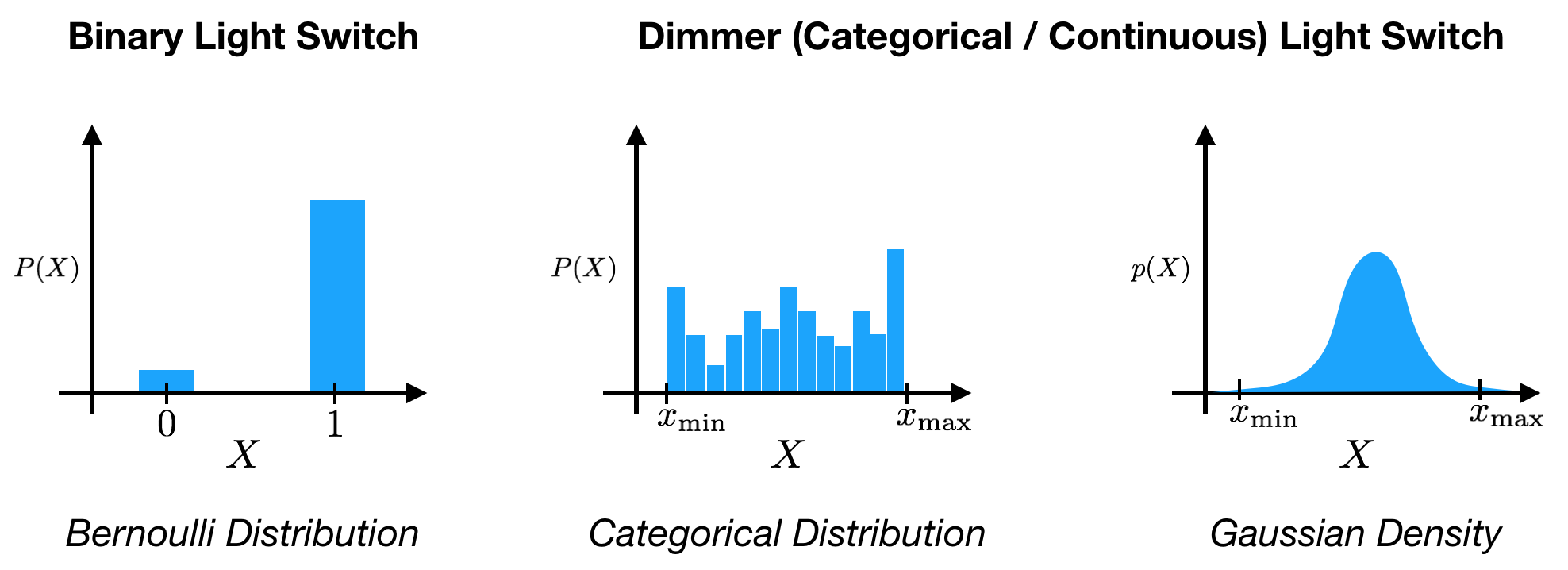
What happens when we swap out our binary light switch for the dimmer switch? The state of the dimmer can be anywhere in the interval from $x_{min}$ to $x_{max}$. We’re confronted with an issue: there are an infinite number of states in this interval. Given that the total probability mass is finite, how do we assign a probability mass to any one of the infinitely many states? The key is the following: even though a variable might have an infinite number of states, our actual observations are never infinitely precise. This arises from limits in our observation capabilities (e.g. hardware sensitivity limits, discrete computer memory) and possibly inherent quantization (e.g. photons of light). Continuous variables are always observed with uncertainty!
So how do we model the probability over the states of a continuous variable? One option is to discretize our continuous space. That is, we break up the observation interval into bins that correspond to our observation uncertainty windows. Then, using a categorical distribution, we assign a probability mass to each of these bins:
\[P(X=x_\ell) = P_\ell \text{ for } \ell=1, \dots, L,\]where we have $L$ bins, with $P_\ell$ the probability for bin $\ell$, and $\sum_\ell P_\ell = 1$. If our observations always fall into the same set of bins, e.g. image pixels take integer values from 0 to 255, then this can be an effective way to model an underlying continuous variable. However, there are drawbacks to this method. When our uncertainty bins are not easily partitioned or if we have a large number of bins, this approach will not work. Likewise, this approach neglects the relational nature of the states, i.e. states are either larger or smaller than others. A categorical distribution would be the same if the bins were arranged randomly.
To overcome these issues, we can use a probability density function, $p(X)$. Note that we’re using $P$ for probability mass functions and $p$ for probability density functions. Rather than assigning a mass to each state, a probability density function specifies a density over states. To find the mass within a window of observations, we integrate the density over this window. As in the case of probability mass functions, we ensure that the probability density must integrate to $1$ because, again, the variable must be in some state:
\[\int p (X) dX = 1.\]Note that the only other requirement for the density is $p(X) \geq 0$, i.e. $p(X)$ can be larger than $1$ at some states. Going back to our dimmer light switch, we could, for example, model the brightness of the dimmer switch as a Gaussian density, $p(X) = \mathcal{N} (X; \mu, \sigma^2)$, specified by a mean, $\mu$, and a variance, $\sigma^2$ (although a Beta density would be more appropriate). The mean is the center of our estimate, and the variance controls the width of that estimate. When we make an observation of the brightness resulting from the dimmer switch, we then take our Gaussian density and integrate over our observation window to find the probability mass assigned to that observation. If our observation is in the window $x_\text{obs} = [ x_\text{obs min}, x_\text{obs max} ]$, then we calculate
\[P(x_\text{obs}) = \int_{x_\text{obs min}}^{x_\text{obs max}} \mathcal{N} (X; \mu, \sigma^2) dX.\]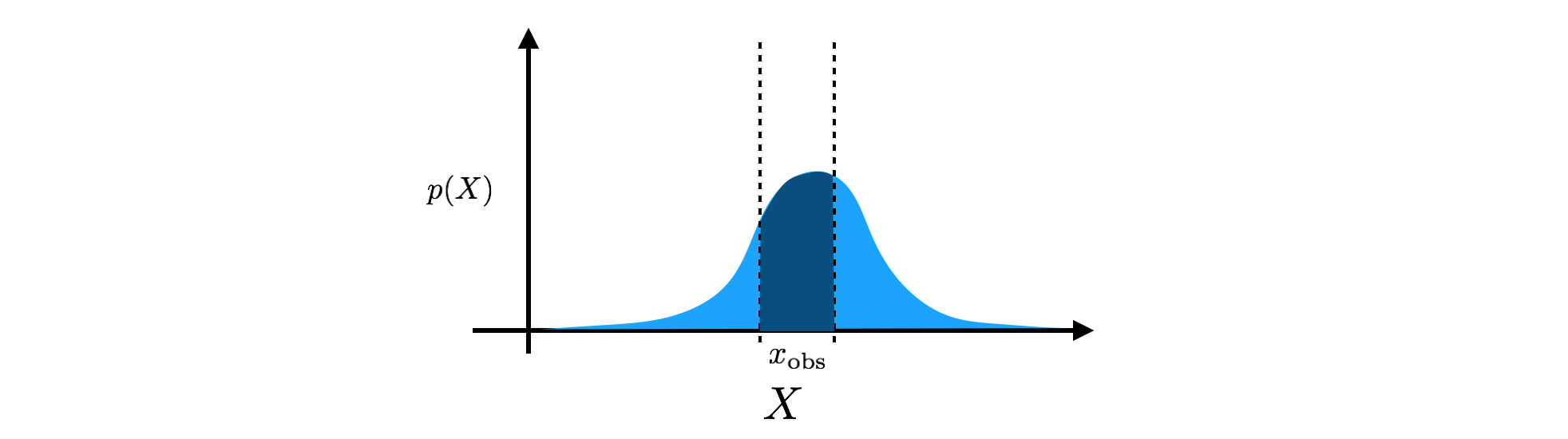
Equivalently, we can use the density’s cumulative distribution function (CDF), which quantifies the amount of probability mass under the curve from $-\infty$ to each point. Calculating the probability mass within a window then becomes a calculation of the difference between the CDF at the edges of the window:
\[P(x_\text{obs}) = \text{CDF} (x_\text{obs max}) - \text{CDF} (x_\text{obs min}).\]Finally, while we have focused on evaluating probability masses, in some cases, e.g., when comparing densities, we may only care about evaluating the density itself at a single point. Note that because $p(X)$ can be larger than $1$, $\log p(X)$ can be positive in these situations.
latent variable models + variational inference
We’re now going to apply these ideas to latent variable models. A latent variable model is a probabilistic graphical model that models an observed variable, $X$, as being generated from an underlying latent variable, $Z$. This is done using parameters, $\theta$, that define a joint probability,
\[p_\theta (X, Z) = p_\theta (X \| Z) p_\theta (Z).\]The joint distribution quantifies the co-occurrence probability of each value of the latent variable with each value of the observed variable. The types of $X$ and $Z$ are distinct; either variable can be binary, discrete, continuous, etc., depending on the data and the assumptions of the model. Note that I have used the lower case $p$ here, however, whether these are distributions or densities depends on the forms of the variables.
Learning the model parameters, $\theta$, or inferring the posterior distribution over the latent variable from an observation, $p (Z | X)$, are both computationally intractable for most model classes. This arises from the fact that both tasks require evaluating the (marginal) likelihood:
\[p_\theta (X) = \int p_\theta (X, Z) dZ,\]which involves integrating over the latent variable. With a continuous $Z$ or a complicated model, this is computationally costly. For this reason, we resort to approximate inference methods. Variational inference introduces an approximate posterior, $q(Z | X)$, that induces a lower bound on the marginal log-likelihood, often referred to as the evidence lower bound (ELBO):
\[\mathcal{L} = \mathbb{E}_{Z \sim q(Z \| X)} \left[ \log p_\theta (X, Z) - \log q (Z \| X) \right] \leq \log p_\theta (X).\]Inference and learning then become alternating optimization problems. During inference, we optimize $\mathcal{L}$ w.r.t. $q (Z | X)$. During learning, we optimize $\mathcal{L}$ w.r.t. $\theta$. This is referred to as the variational EM algorithm. We can equivalently write the ELBO as
\[\mathcal{L} = \mathbb{E}_{Z \sim q(Z \| X)} \left[ \log p_\theta (X \| Z) \right] - D_{KL} (q (Z \| X) || p_\theta (Z)),\]where $D_{KL}$ denotes the KL-divergence between the two distributions within the parentheses. The first term in this expression, the conditional log-likelihood, quantifies how well the model fits (reconstructs) the observation. The second term quantifies the agreement between the approximate posterior and the prior, which can be considered as a form of regularization. Note that because $\mathcal{L}$ is a bound on $\log p_\theta (X)$, our discussion about probabilities above also applies to the ELBO.
Latent Gaussian models use continuous latent variables with Gaussian densities, i.e. $p_\theta (Z) = \mathcal{N} (Z; \mu_p, \sigma_p^2)$. Again, $p_\theta (X | Z)$ can take any form that can model the observations. The original papers that introduced variational auto-encoders (VAEs) (here and here) presented quantitative results using Bernoulli output distributions on the binarized MNIST data set. Evaluating $\log p_\theta (X | Z)$ is straightforward in this case: we simply evaluate how much probability mass the model placed at the observed binary values. The papers also present results using Gaussian output densities on natural images. While Rezende et al. only present qualitative results in this setting, Kingma and Welling report ELBO values by evaluating the Gaussian conditional likelihood density at the pixel values.
A number of papers have demonstrated improvements for generation, inference, or both. For instance, DRAW uses recurrent networks in the inference and generative models. The Ladder VAE adds a “top-down” component to the inference model. Many works have also attempted to bring VAE-type models to the sequential setting, such as VRNN, SRNN, etc., where speech, music, and handwriting datasets have been modeled. Recent works have also looked at using similar models for video data, such as DVBF, KVAE, SVG, etc.
Many of these papers use different evaluation schemes for continuous data. DRAW and SVG evaluate mean squared error (MSE) for the conditional likelihood on natural images. Within a constant, this is equivalent to a factorized Gaussian log-likelihood with a variance of $1$. SVG also averages over spatial dimensions (see the PyTorch documentation on MSE) rather than summing the log probabilities. Other works, such as VRNN, SRNN, Ladder VAE, etc., evaluate the log-density of the output Gaussian at the data points. Finally, some papers, like inverse autoregressive flow, evaluate the probability mass under output densities by effectively integrating over pixel bins.
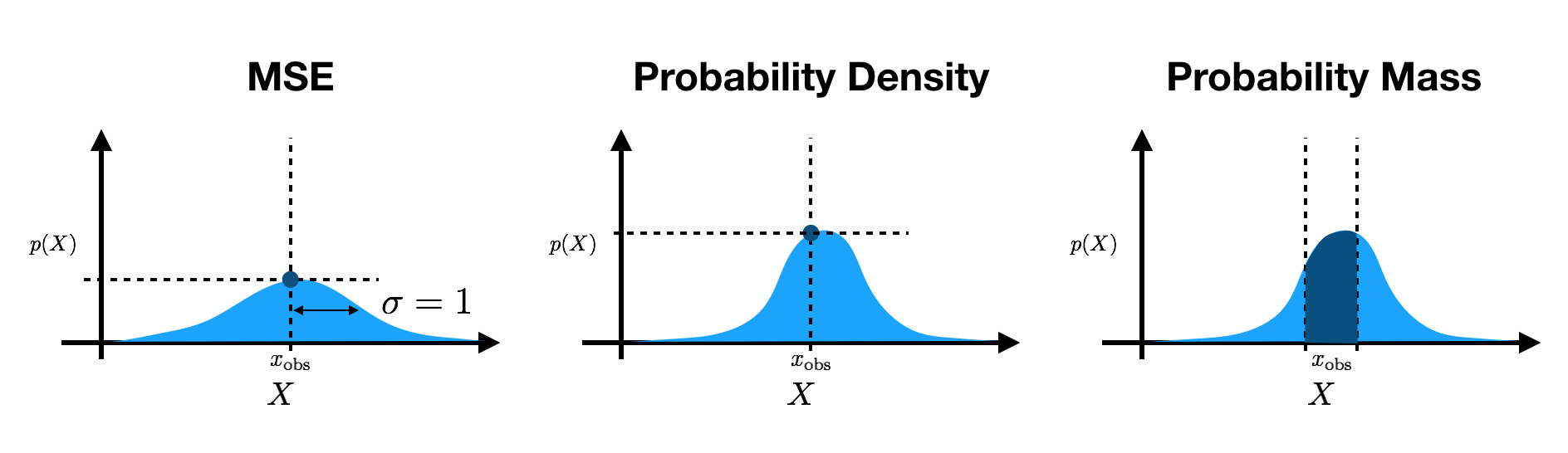
Why does the disparity in evaluation schemes matter? In short, it is important that we be consistent in reporting results. This enables us to quantitatively compare our results across different models that make different assumptions. If we use inconsistent evaluations, the scales (and signs) of reported values will be drastically different, thereby hindering our ability to compare methods and limiting our ability to progress as a field.
modeling continuous observations
To close, let’s walk through the math and code for evaluating probability masses and densities from probability density functions. As our example, we’ll use Gaussian densities, but the same rules apply to other probability densities. The probability density function for a univariate Gaussian is expressed mathematically as:
\[\mathcal{N} (X; \mu, \sigma^2) = \frac{1}{\sqrt{2\pi \sigma^2}} \exp \left( -\frac{(X-\mu)^2}{2 \sigma^2} \right)\]Most of the deep learning libraries provide all of the necessary primitives to implement this density, or more commonly, its logarithm. Many libraries, including Tensorflow and PyTorch, even have all of these probabilistic programming functionalities built directly into the library. For instance, in PyTorch, one can express the log density as:
Again, to evaluate the probability mass within a window, we integrate the density over that window or use the CDF. The CDF for a Gaussian density takes the following form:
\[\Phi (X) = \frac{1}{2} \left[ 1 + \text{erf} \left( \frac{X - \mu}{\sqrt{2 \sigma^2}} \right) \right],\]where $\text{erf}$ denotes the error function. Again, most deep learning libraries provide the primitives to implement this from scratch, or even provide it directly. In PyTorch, this is expressed as:
So, if we want to evaluate, for example, the probability mass under a Gaussian density in one pixel, we need to evaluate the CDF at each end of the pixel window. Often, to make the optimization easier, we rescale images to the interval $[0, 1)$, so the pixel width is effectively scaled to $\frac{1}{256}$. Thus, computing the probability mass of an image, $x_\text{image}$, is as straightforward as calculating the CDF at $x_\text{image} + \frac{1}{256}$ and subtracting the result of the CDF at $x_\text{image}$:
\[P(x_\text{image}) = \text{CDF} (x_\text{image} + \frac{1}{256}) - \text{CDF} (x_\text{image}).\]And with that, we can evaluate the probability mass assigned by our model.
some extra notes
Some auto-regressive models, such as Pixel RNN, use categorical outputs to model images. Convolutional DRAW uses a different technique for evaluating Gaussian log-likelihoods, based on comparing the ratio between the density and a uniform density. Pixel CNN++ uses a discretized Logistic density to model images. However, perhaps the most exciting development in the area of density estimation is the use of invertible transforms that allow one to learn intermediate spaces where it is easier to model the data distribution. An example of this is Real NVP, but the same idea is used in Normalizing Flows. See Eric Jang’s blog posts for a nice discussion of this technique.
